-
1. What is HLMA?
The
HLMA
is a curated database for lung microbes across the upper and lower respiratory tract and their
associations with health and respiratory diseases (e.g., COPD, asthma and LUAD). The development
of HLMA aims to: (i) to promote the reusability of metagenomic data (e.g., mNGS, 16S rRNA and ITS)
associated with human lung microbiome (e.g., archaea, bacteria, viruses and fungi); (ii) to assist
users to browse or search the abundance and prevalence of lung microbes across different body
sites and different disease types; (ii) To facilitate the identification of
disease-related/phenotype-related lung microbes.
The HLMA database encompasses the following features:
-
Browse or search microbes across upper and lower respiratory tract and their associations with
health and respiratory diseases (including COPD, LUSC, LUAD, etc.);
-
Browse or search the composition and diversity of the microbiome between groups in each
independent dataset;
- Explore potential microbes associated with respiratory diseases;
-
Perform LAD score analysis on microbiome by submitting a new matrix containing of the abundance
of microbes, along with the corresponding meta-data for each sample;
- Download all results and figures for further research.
-
2. What is lung microbiota and how can it be identified and quantified?
A growing body of evidence indicates that lung microbiome is emerging as potential contributors to
chronic lung diseases, even lung cancer. The lung microbiota is the set of archaea, bacteria,
fungi viruses and protozoa, which reside in the lower airways. The oral and upper respiratory
tract microbes shape the lung microbiota. So, in order to achieve a comprehensive understanding of
the composition and characteristics of lung microbiota, thousands of specimens from six body
sites, covering the upper and lower respiratory tract, were integrated in the HLMA database.
With the advancement of sequencing technologies, metagenome sequencing technology (e.g., mNGS, 16S
rRNA and ITS) is used by researchers to parse complex microbial communities and their functional
capabilities. So far, we collected and curated xx metagenomic datasets from
2,578
metagenomic datasets from multiple public databases in HLMA, which contained
xx
samples and belonged to health and
38
diseases. The counts/abundance of each microbe per sample was quantified by using amplicon
sequence variants (Qiime2 for 16S/ITS) or k-mer-based approach (Kraken2 for mNGS).
-
3. Data processing and quality control
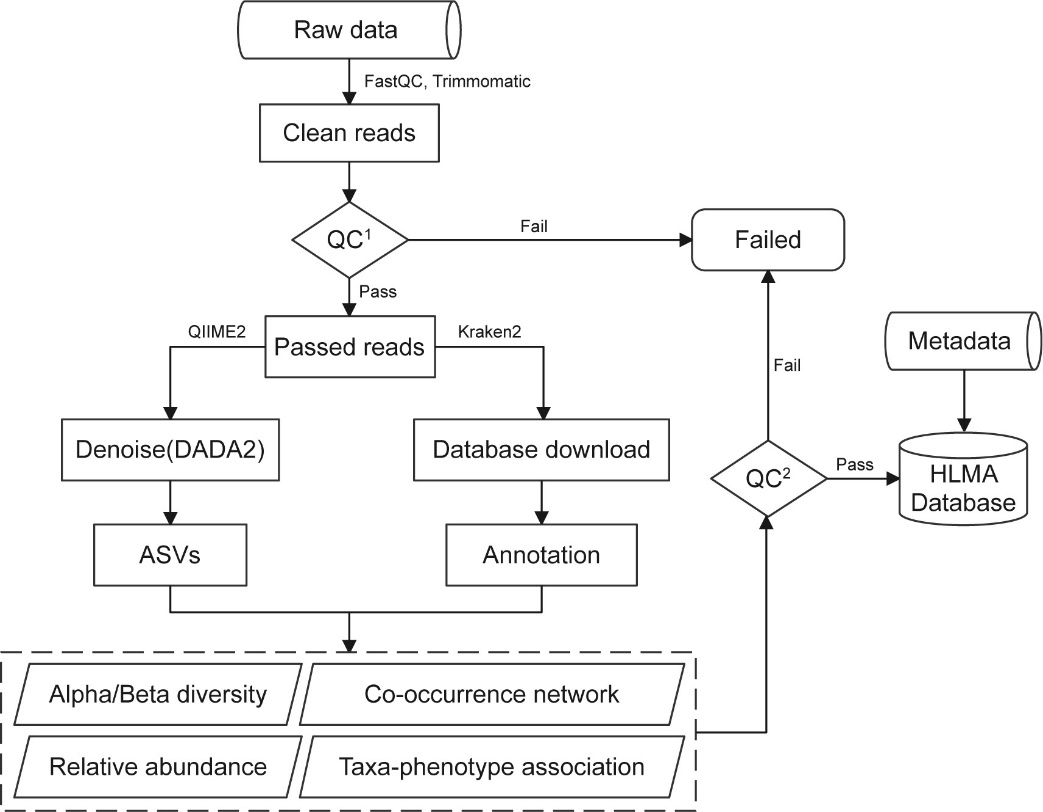
The overall workflow of data processing is depicted in
Fig.1,
including a series of bioinformatic analysis (e.g., adapters trimming, low-quality reads removing,
taxonomic assignment and relative abundance estimation). In HLMA, to ensure data quality, the
following criteria were applied for all samples/runs: (i) samples/runs with fewer than 5,000 reads
were excluded; and (ii) samples/runs in which a single species or genus that accounted for 99.99%
or more of the total abundance were excluded.
Fig. 1 The overall workflow of data processing in HLMA
-
4. Database construction and web development
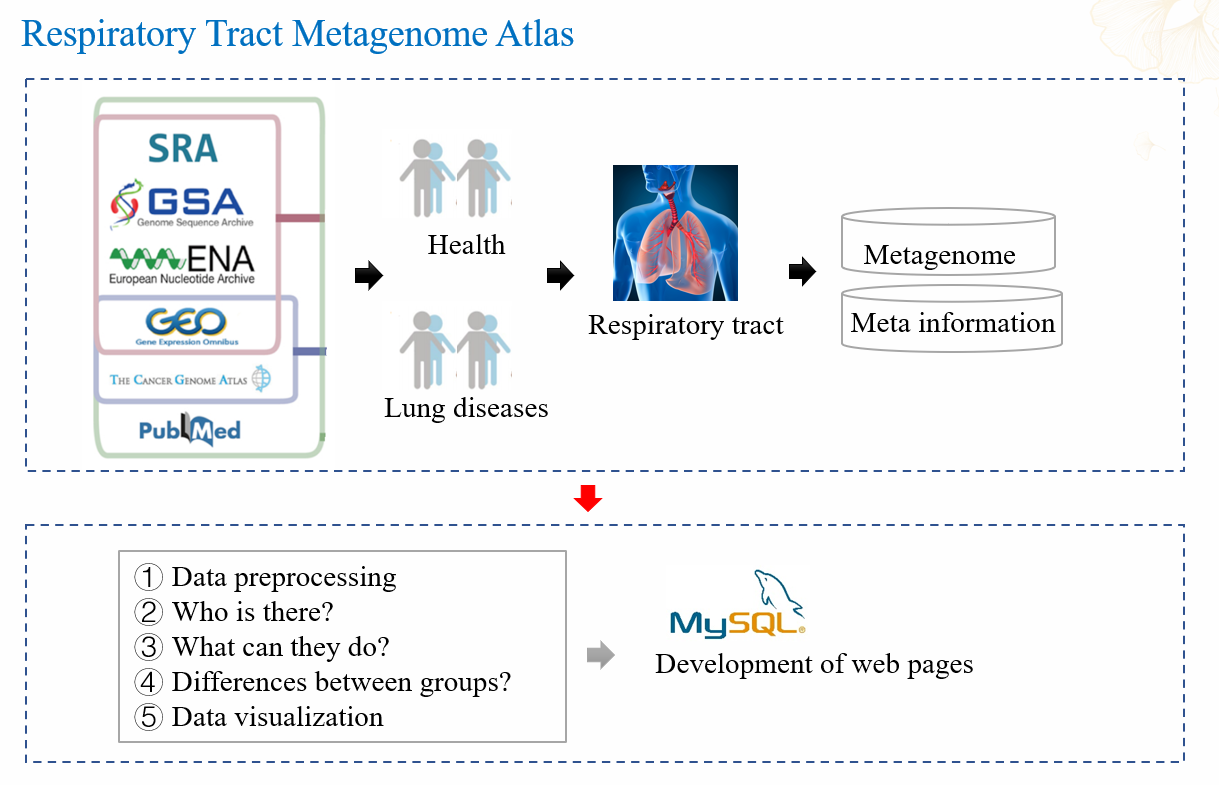
The overall framework for HLMA development is illustrated in
Fig. 2.
All data in HLMA stored into the MySQL (v5.7.25) database. The development of the front-end
webpages utilized HTML5 and JavaScript, while Java (v1.8) with the Spring framework (v1.1.2) for
back-end webpages. We utilized jQuery (v1.8.3) and Bootstrap (v5.3.0) to bridge the front-end and
back-end components, along with other open-source libraries such as echarts.js (v5.4). The website
was hosted on a nginx server (v1.23.3).
Fig. 2 The development of HLMA
-
5. How to query the database?
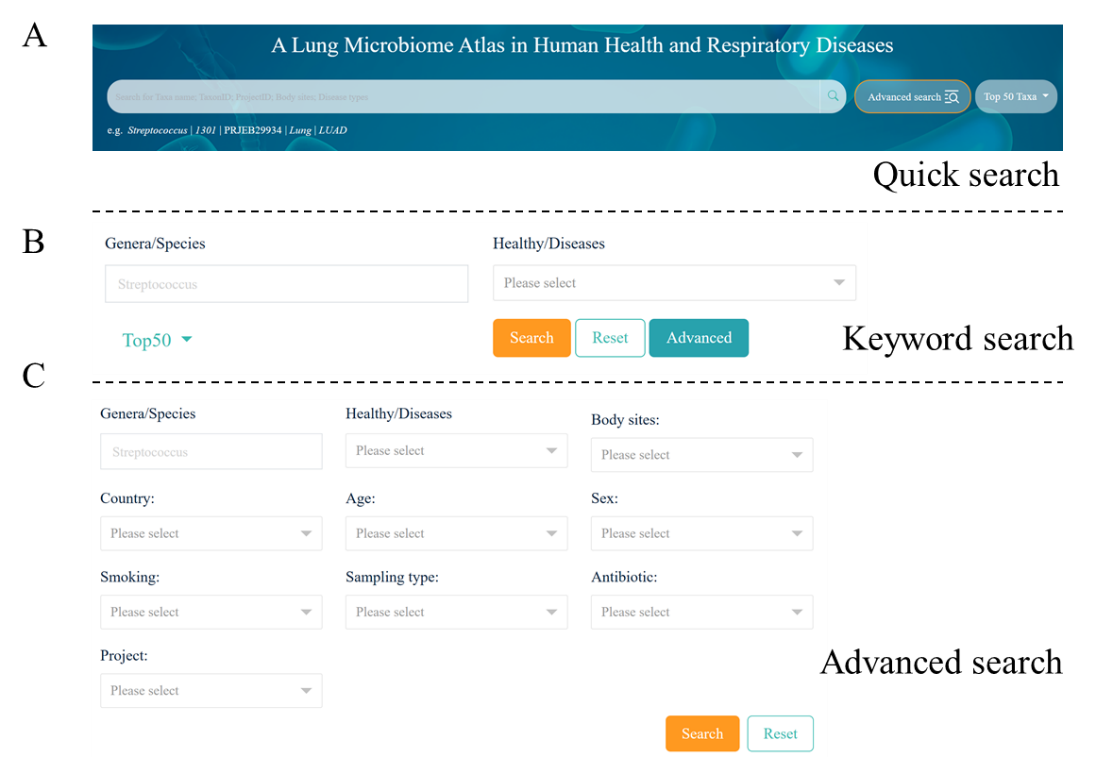
HLMA is a user-friendly web interface that provides to allow users to query the database from
three distinct web pages, including homepage, search page and advanced search page, with multiple
criteria, such as sampling type, genus/species, disease type, gender or age
(Fig. 3A-C).
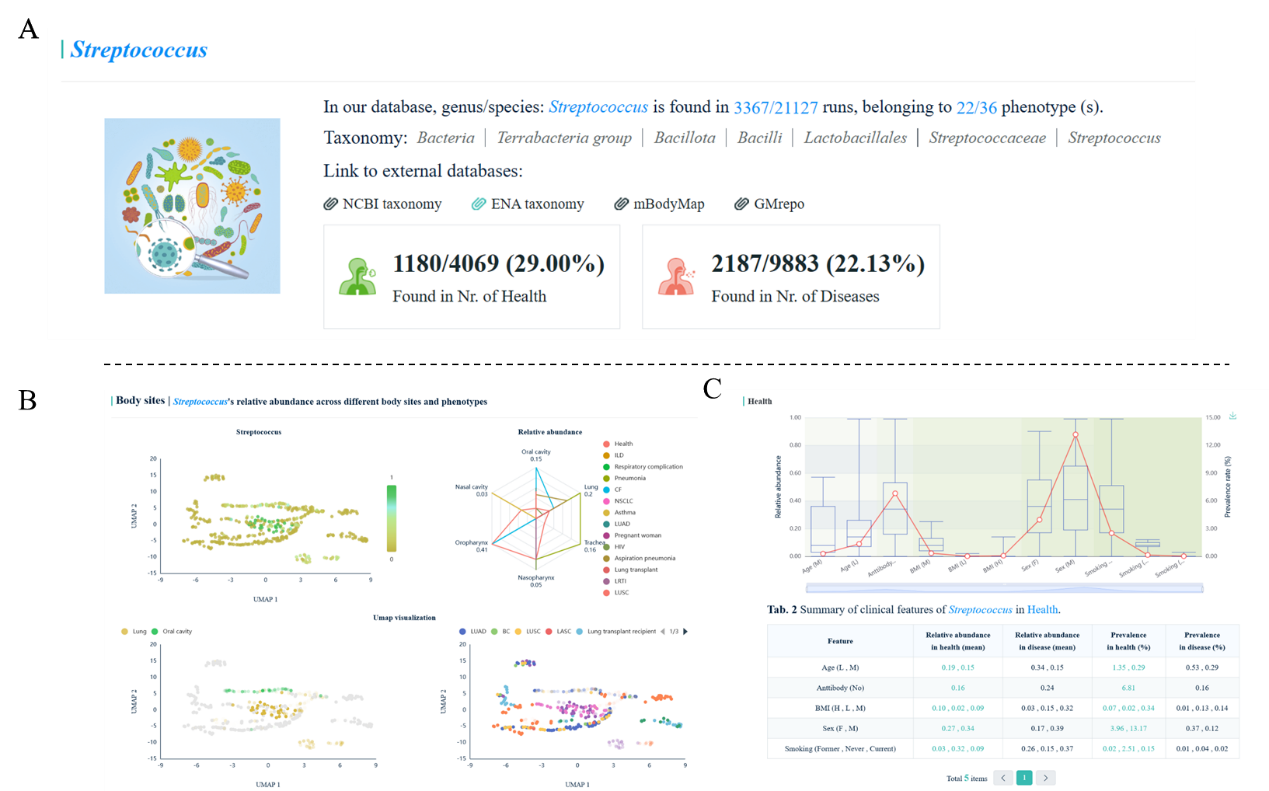
Once quiring one genus/species, all relevant results are visualized, including the general
description for the taxa
(Fig. 4A),
the relative abundance of prevalent of the taxa across various body sites
(Fig. 4B),
health and different respiratory diseases
(Fig. 4C),
as well as in different regions/countries
(Fig. 4D).
It is worth mentioning that the relative abundance of the taxa across different body sites was
visualized using UMAP in the section "Body sites"
(Fig. 4B).
Additionally, the prevalence of the taxa was visualized using a global map, providing a visual
representation of its distribution across different regions worldwide. This visualization not
only highlights the overall prevalence but also allows for the exploration of its distribution
among different diseases within each region
(Fig. 4D).
A detailed table is available below each figure, which presents detailed statistics of the taxa
under different conditions. To enhance user convenience and accessibility, each table is
equipped with functional features, including sort for certain column, download the table in TSV
(Tab-Separated Values) format, and search by using a keyword.
Fig. 3
Search function in HLMA, (A) Quick search across global database in the "Home" page by
a keyword, such as Streptococcus, LUAD, and lung; (B) Keyword search in the "Search" page by
using one and/or two criteria; (C) Advanced search in the "Advanced search" page by using
multiple criteria, such as sampling type, genus/species, disease type, gender or age.
Fig.4
Taking the genus Streptococcus as an example and visualization of all relevant
results. (A) The general description of the taxa, such as taxonomy and links to external
databases; (B-D) The relative abundance of prevalent of the taxa across various body sites
(B), health and different respiratory diseases (C), as well as in different regions/countries
(D).
-
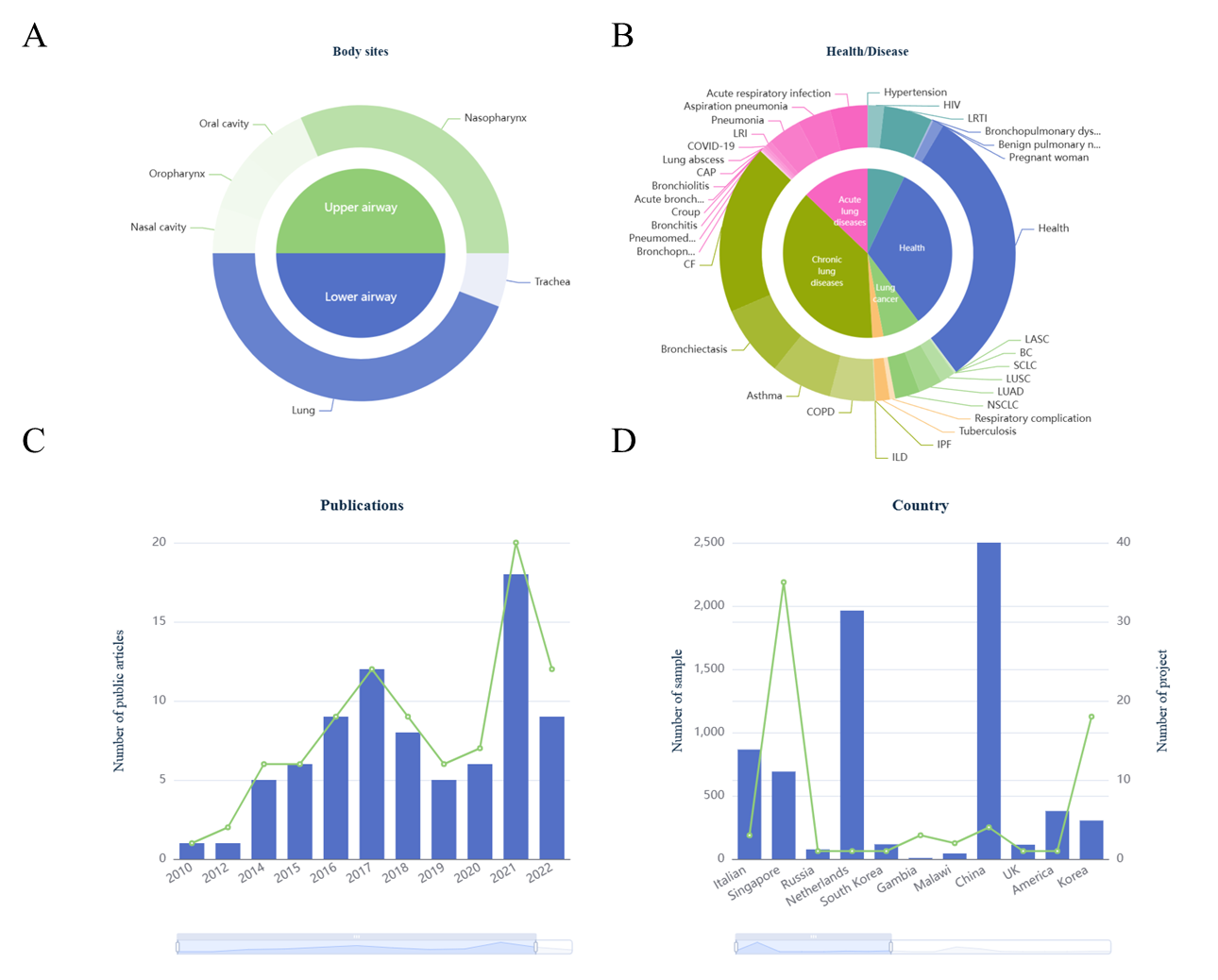
6. Data overview and statistics.
Currently, both the "Home" page and the "Statistics" page in HLMA display the distribution of
manually collected datasets as well as related metadata. A total of
21,833
runs/samples from
92
independent datasets, covering the upper and lower respiratory tract, including
7 body sites
(Fig.5A),
were curated in HLMA (Fig.5).
Fig.5
The pie chart illustrating project and sample sizes across
different body sites (A) and various health/diseases (B) as well as barplots depicting the
distribution of publication per year (C) and by region/country (D).
-
7. How to use toolkit?
Currently, three toolkits were embedded in HLMA, including Summary of key studies,
Drug resistance analysis, and submission. The tutorial of those toolkits as follows:
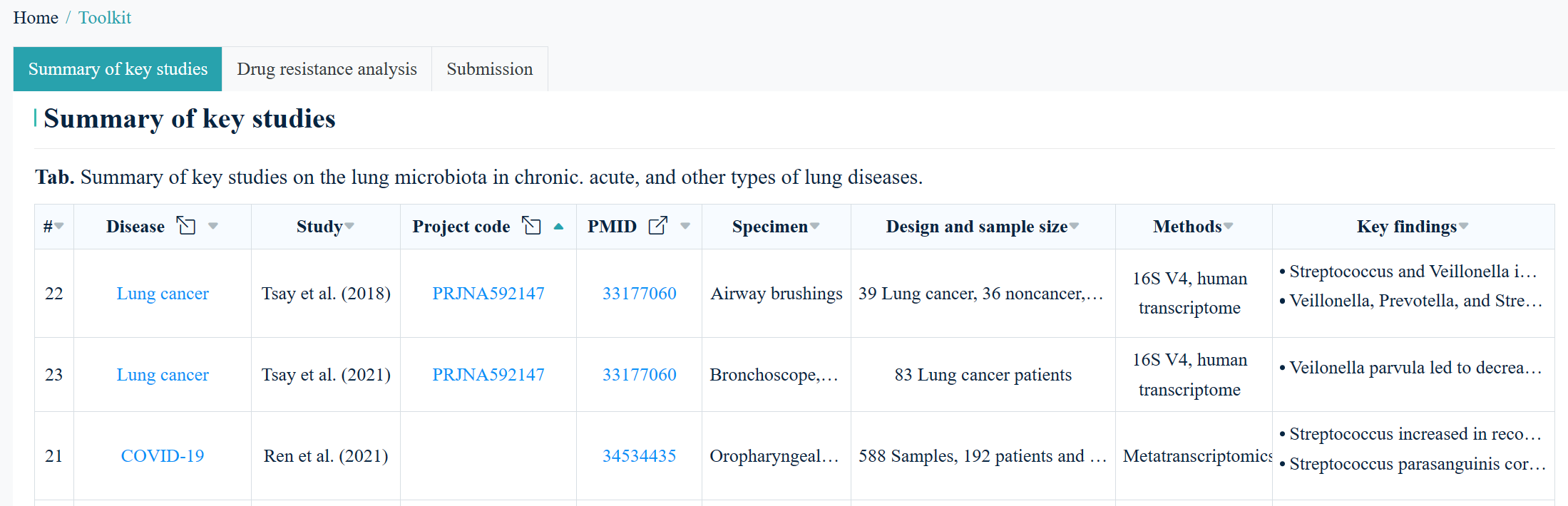
7.1 Summary of key studies
This section displays current knowledge of the lung microbiome in human diseases, spanning
across
chronic, acute, and other types of lung diseases
(Fig. 6).
If a dataset is available, users can click on it to access a more detailed page, just like on
the
"Search" or "Browse" page.
Fig.6
Summary of key studies on the lung microbiota in chronic,
acute, and other lung diseases.
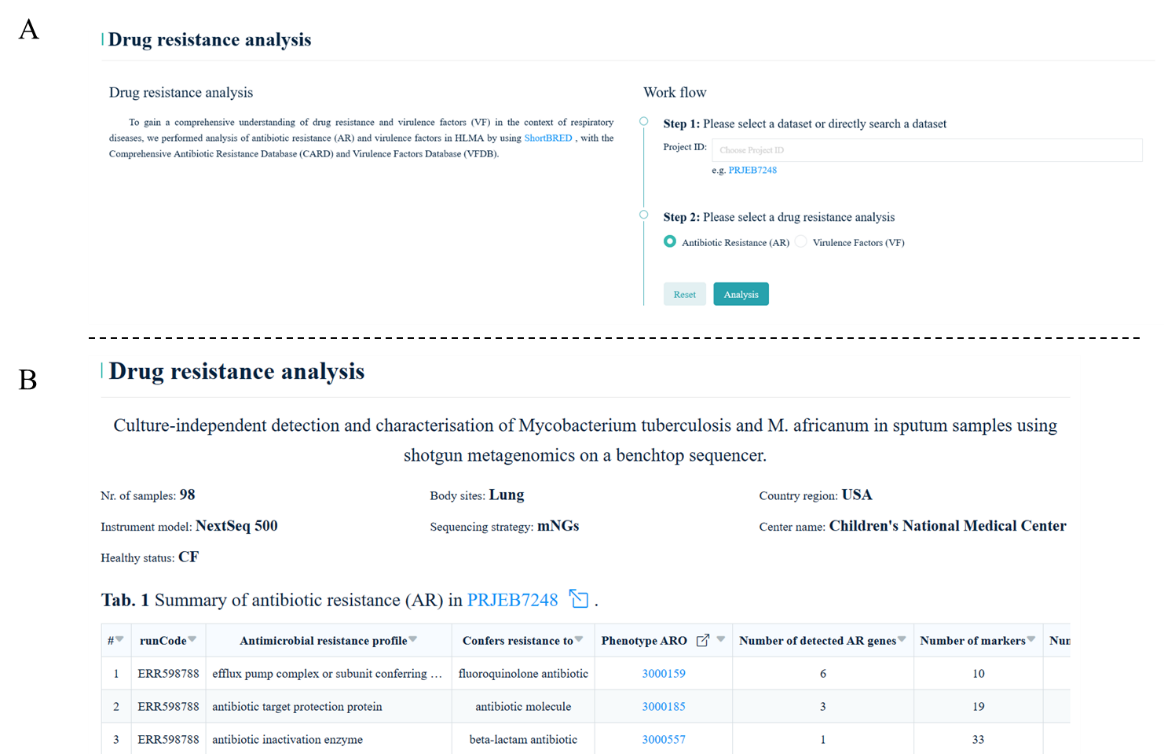
7.2 Drug resistance analysis
The drug resistance analysis is a helpful application to discover and identify the potential
antibiotic resistance (AR) and virulence factors (VF) by using mNGS data. Once a special dataset
the user chooses, the results are displayed as table format
(Fig. 7).
Fig.7
Taking
PRJNA7248
as an example to display the results of drug resistance analysis.
7.3 Differential abundance test
The tool enables users to submit their new data for detection of differential
abundance taxa. We used R package microeco (version 0.20.0) to identify condition-specific
taxa
(Fig. 8).
On the "Differential abundance test" page, users firstly fill in some basic
information about project
(Fig. 8A).
Then, some important parameters will need to set by users before
executing the program, such as method (e.g., LEfSe, ANOVA), taxonomic level (e.g.,
genus,class)
(Fig. 8B).
Once a click on "Run", the related details will send to them in the format of an
Email. The "*" symbol indicates the required fields.
Fig.8
The toolkit "Differential abundance test" page in HLMA.
-
8. What's ChartDoc?
To enhance user experience, ChatDoctor (Cite as arXiv:2303.14070;
https://arxiv.org/abs/2303.14070), a specialized medical chat model, is embedded in the upper
right corner of the page. Once querying a keyword or one question, user can access detailed taxa
and other related description anytime, day or night. This can be particularly beneficial for
individuals seeking immediate information.